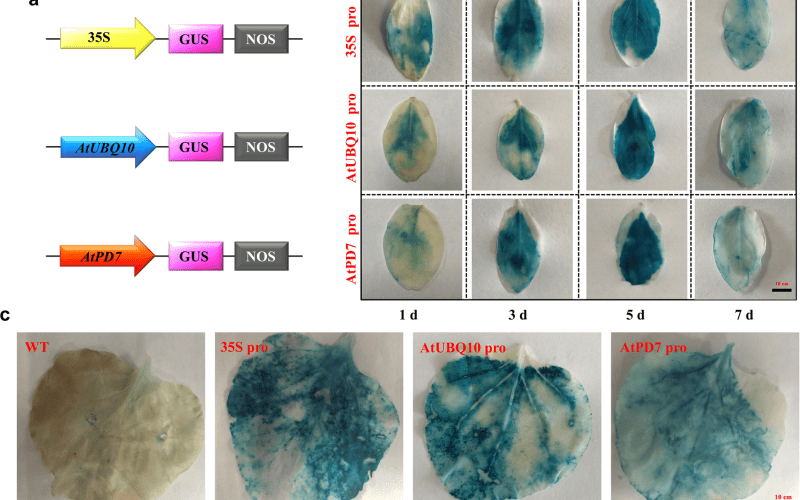
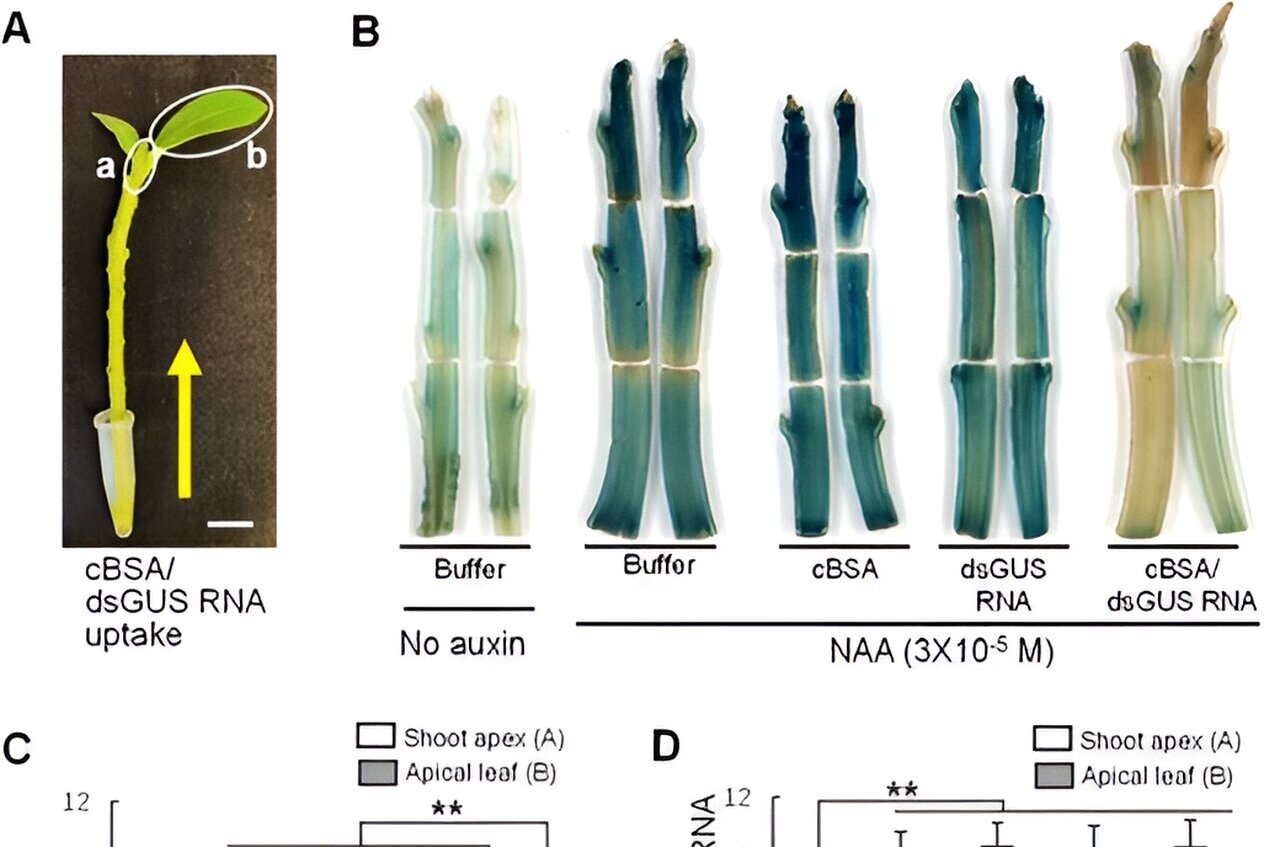
The GUS assay, utilizing the enzyme β-glucuronidase, has long been a trusted method in plant biology for monitoring gene expression. Its adaptability across various plant species, along with high sensitivity and clear spatial resolution, has contributed to its widespread acceptance.
However, the interpretation of GUS assay data can sometimes be overly simplistic, leading researchers to draw misleading conclusions, particularly in studies that focus on developmental regulation, stress responses, or nuanced expression differences.
This article aims to re-examine the GUS assay from both an interpretative and experimental design standpoint, emphasizing how biological context, assay characteristics, and experimental conditions can shape the understanding of GUS reporter signals.
Understanding the GUS Signal: More Than Just Transcription
Ultimately, a GUS signal is a representation of the accumulated enzymatic activity of β-glucuronidase rather than an immediate reflection of transcription levels. This distinction is crucial for accurate interpretation.
Various factors, including the stability of GUS proteins, the turnover rate of the enzyme, and the availability of substrates, all contribute to the overall signal intensity. Thus, GUS staining does not merely indicate current transcriptional activity; rather, it reflects a time-averaged expression of the target gene.
In tissues that undergo rapid cell division or during key developmental transitions, the integration of gene expression over time can blur the visibility of transient promoter activation. Conversely, detectable signals may linger even after transcription has terminated due to the stability of GUS protein. Recognizing this temporal disconnect is vital, especially when studying intricate regulatory processes where timing plays a crucial role.
Examining Spatial Resolution in GUS Assays
One of the standout features of the GUS assay is its capacity to highlight spatial expression patterns within plant tissues. However, this spatial resolution can be significantly affected by the physical and biochemical attributes of the plant material. Factors such as the integrity of cell walls, layers of cuticle, and tissue density can greatly influence how well the staining solution penetrates and interacts with tissues.
As such, a lack of staining in certain regions doesn’t automatically signal an absence of gene expression. Instead, it might merely indicate differential permeability of the staining solution, which can skew the perceived specificity of expression. Therefore, it is essential for researchers to interpret spatial data with an understanding of the underlying tissue architecture while also validating findings through complementary methodologies.
Comparing Expression Strength with Fidelity
In practice, researchers frequently assess promoter activity by comparing the intensity of GUS staining. However, this approach can lead to complications. High levels of expression may overwhelm the enzymatic reaction, causing the signals to diffuse into surrounding areas and misleadingly suggest a broader expression pattern than is actually present.
On the flip side, tighter regulation from weaker promoters might yield accurate tissue-specific signals that are harder to detect. Although extending staining durations or increasing substrate concentrations could enhance visibility, doing so may also introduce background noise, complicating analyses. Thus, achieving a balance between visibility and fidelity is a key challenge in GUS-based studies.
The Influence of Developmental Context
The developmental stage of plant tissues has a profound impact on both GUS expression and the performance of the assay itself. Different phases of development are associated with variations in cellular metabolism, vacuolar pH, and redox states, all of which can affect GUS enzyme activity independently of transcriptional control. Therefore, seedlings, meristems, and aging tissues could display vastly different staining results even when analyzed under the same promoter control.
Failure to account for these developmental differences may result in erroneous conclusions about gene expression related to specific life stages. For longitudinal studies, maintaining consistent sampling regimes and parallel control groups is essential to delineate between genuine regulatory changes and developmental influences.
Environmental Variables and Stress Impact
Environmental factors introduce additional complexities to GUS assays. Fluctuations in temperature, variations in light exposure, and the presence of stressors can significantly affect not just promoter activity but also the stability and efficacy of the enzyme involved. In stress-response investigations, GUS expression patterns might mirror cellular stress states that alter the behavior of the reporter rather than reflecting transcriptional regulation alone.
This consideration is especially crucial when using GUS assays to assess stress-induced promoters. Without careful controls and well-defined conditions, researchers may find it challenging to distinguish true transcriptional activation from stress-related adaptations in reporter behavior.
Leveraging GUS Assays: Strengths and Limitations
Given the constraints inherent in GUS assays, they are most effective when used in comparative frameworks. Evaluating differences between constructs, treatment groups, or genotypes processed simultaneously tends to yield more insightful data than relying on absolute measures of signal intensity.
Cross-study comparisons can be particularly problematic due to variations in staining protocols, incubation times, and tissue handling methods. Researchers need to exercise caution when interpreting quantitative differences unless strict standardization across assays has been maintained.
Integrating GUS Assessments with Other Expression Techniques
To improve the clarity and interpretive power of GUS assays, it is increasingly beneficial to integrate them with other expression analysis techniques. For instance, quantitative PCR can provide more nuanced transcriptional insights, while fluorescent reporters can offer real-time monitoring and subcellular localization of gene expression. Additionally, simultaneous analysis of endogenous gene expression can ground reporter data within a more physiological context.
In this light, GUS assays should be viewed not as standalone measurements but rather as components of a well-rounded research strategy that encompasses spatial, temporal, and quantitative dimensions.
Avoiding Common Misinterpretations
A prevalent source of error in studies employing GUS assays arises from equating staining intensity with promoter strength across unrelated systems. Another misleading assumption is that a lack of signal signifies the absence of expression. Both misinterpretations overlook the inherent biological and technical variables associated with enzymatic reporter systems.
Acknowledging these limitations does not diminish the importance of the GUS assay; rather, understanding them enables researchers to design experiments more effectively and interpret data more accurately.
Conclusion
The GUS assay remains an invaluable tool in plant research, but its interpretative power hinges on a comprehensive understanding of biological context and assay-specific properties. GUS signals encapsulate integrated enzymatic activity shaped by developmental context, tissue accessibility, and environmental conditions.
When utilized as a comparative tool within a broader research strategy, and paired with complementary analyses, the GUS assay continues to provide profound insights into plant gene expression beyond mere signal detection. Understanding and addressing its limitations will further enhance the quality and relevance of findings derived from this methodology.