What is ChIP Sequencing?
Chromatin immunoprecipitation (ChIP) is an effective method for isolating DNA fragments bound to specific proteins in vivo. DNA-binding proteins can be transcription factors (TFs) or other chromatin-associated proteins, such as histones. With the rapid development of DNA sequencing technology, the direct sequencing of DNA fragments obtained in ChIP using high-throughput sequencing is called ChIP-seq. Due to its high resolution and coverage, ChIP-seq has become a popular method for analyzing chromatin modifications and TF binding sites in eukaryotic genomes.
Promoters and Enhancers: Key Regulatory Elements in Gene Expression
The genome is composed of both coding and non-coding DNA sequences, with the latter playing a crucial role in regulating gene expression. Among the most important non-coding regulatory elements are promoters and enhancers, which work in concert to control when, where, and how strongly a gene is transcribed.
Promoters: The Gatekeepers of Transcription Initiation
A promoter is a specific DNA sequence located near the transcription start site (TSS) of a gene, serving as the binding site for RNA polymerase and other essential transcription machinery. Promoters contain highly conserved sequence motifs, such as the TATA box (found in many eukaryotic genes) and the initiator (Inr) element, which help recruit transcription factors (TFs) and RNA polymerase II to initiate transcription. The strength and specificity of a promoter determine how efficiently a gene is transcribed. Some promoters are constitutive, driving constant expression, while others are inducible, responding to environmental or cellular signals.
In addition to the core promoter region, which directly interacts with RNA polymerase, many genes also possess proximal promoter elements located within a few hundred base pairs upstream of the TSS. These sequences bind transcription factors that either enhance or suppress transcription, fine-tuning gene expression levels.
Enhancers: Long-Distance Transcriptional Amplifiers
Unlike promoters, enhancers are regulatory DNA sequences that can be located far away—sometimes thousands or even millions of base pairs—from their target genes. They function by looping in three-dimensional space to physically interact with promoters, significantly boosting transcription levels. Enhancers are highly flexible in their positioning and orientation; they can be upstream, downstream, or even within introns of the genes they regulate, and they can function bidirectionally.
Enhancers are characterized by specific epigenetic signatures, including:
- Histone modifications such as H3K4me1 (a mark of enhancer regions) and H3K27ac (indicating active enhancers).
- Binding of transcription factors and coactivators like p300/CBP, which help stabilize the enhancer-promoter interaction.
- Open chromatin structure, making them accessible to regulatory proteins.
Enhancers play a critical role in cell-type-specific gene expression, allowing the same gene to be activated in certain tissues while remaining silent in others. For example, enhancers regulate developmental genes, immune responses, and neuronal functions by integrating signals from multiple transcription factors.
Key Differences Between Promoters and Enhancers
- Positional Flexibility: Promoters are fixed near the TSS, whereas enhancers can act over long distances and in any orientation.
- Mechanism of Action: Promoters directly recruit RNA polymerase, while enhancers modulate promoter activity through chromatin looping.
- Dependency: Enhancers can influence multiple promoters, whereas promoters typically regulate a single gene.
Recent studies have also identified super-enhancers—clusters of enhancers that drive high-level expression of genes critical for cell identity, such as those involved in stem cell pluripotency or cancer progression. Understanding these regulatory elements provides insights into gene regulation mechanisms and offers potential therapeutic targets for diseases caused by dysregulated gene expression.
In summary, promoters and enhancers are fundamental to transcriptional control, with promoters serving as the entry point for transcription machinery and enhancers acting as powerful modulators that ensure precise and dynamic gene expression across different biological contexts.
ChIP-Seq Distinguishes Enhancers and Promoters by Analyzing Histone Modifications
Histone modifications generally affect transcriptional activity by affecting the affinity of histones to DNA duplexes, thereby altering the nucleosome structure and the sparse or condensed state of chromatin, or by affecting the affinity of other transcription factors to structural gene promoters to exert a regulatory effect on gene expression. Since histone modifications alter the sparse or condensed state of chromatin, the accessibility of chromatin is also altered.
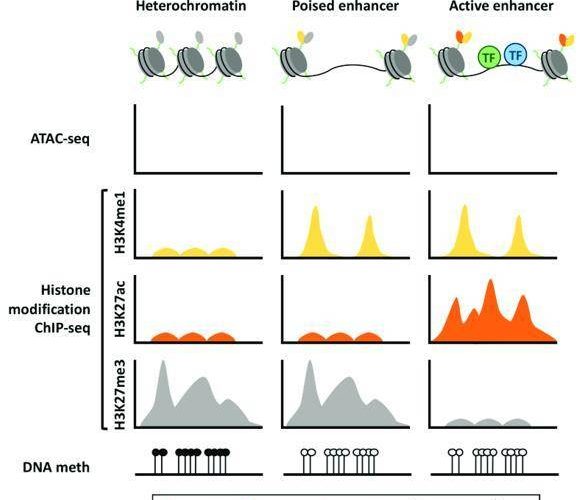
Thus, histone modifications can predict the type of chromatin (heterochromatin or euchromatin), distinguish functional elements of the genome (promoters, enhancers, gene bodies) and detect the decision to have these elements in an active or repressed state. For example, H3K4me2 and H3K4me3 modifications are mostly enriched in promoters near the transcription start site to activate gene expression, while H3K27me2 and H3K27me3 are associated with gene repression.

Chromatin landscape for heterochromatin, poised and active enhancer regions (Ordoñez R et al. 2019)
Therefore, the distribution of histone modifications can be analyzed by ChIP-seq to find the promoter and enhancer regions of genes and whether they activate or repress gene expression.
Common Histone Modifications
Five core histone modifications are widely used for ChIP-seq analysis:
- H3 lysine 4 monomethylation (H3K4me1) or H3 lysine 27 acetylation (H3K27ac), associated with enhancer regions
- H3 lysine 4 trimethylation (H3K4me3), associated with the promoter region
- H3 lysine 36 trimethylation (H3K36me3), associated with the transcriptional region in the gene body
- H3 lysine 27 trimethylation (H3K27me3), associated with Polycomb repression
- H3 lysine 9 trimethylation (H3K9me3), associated with heterochromatin
Application of Super-Enhancer Research
When multiple enhancers are clustered in certain regions while being densely bound by a series of transcription factor proteins, they regulate the activation, high-level expression of a large number of cell-type-determining associated genes, and this region is called a super-enhancer (SEs). Super-enhancers can strongly drive the expression of genes controlling cell identity and can be used to explain cell type-specific expression patterns, showing great potential for application in the study of the pathogenesis of diseases, such as developmental biology and cancer. Super enhancers are usually identified by ChIP-seq, and screening of key SEs and their regulatory genes is important for elucidating the regulatory mechanisms of key driver genes in diseases and tumors, anti-tumor function studies, and drug screening.
learn more: ribosome profiling
Reference:
- Ordoñez R, Martínez-Calle N, Agirre X, et al. DNA methylation of enhancer elements in myeloid neoplasms: think outside the promoters? Cancers, 2019, 11(10): 1424.