Introduction
Cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA) are both valuable biomarkers in liquid biopsies, but they differ in origin, biological significance, and clinical applications. Understanding these differences is crucial for selecting the right extraction and analysis methods for diagnostic, prognostic, and monitoring purposes. This guide explains their biological differences, optimal extraction methods, and real-world applications-with practical tips for researchers and clinicians.
Comparative Overview of cfDNA and ctDNA
1. Fundamental Differences Between cfDNA and ctDNA
cfDNA and ctDNA come from different places in the body and have different roles.
- cfDNA mainly comes from normal cells that are dying or being broken apart. It’s mostly made up of 166 base pairs, thanks to the protection from the cell parts that hold the DNA together. In healthy people, cfDNA is found in plasma at levels between 1-100 ng/mL, and it doesn’t usually have any changes in the genes. Because of this, cfDNA is great for checking how the body’s cells are working in general.
- On the other hand, ctDNA comes from cancer cells. These cells release ctDNA when they die, break apart, or actively let it go into the bloodstream. ctDNA has two types of pieces: short ones (less than 150 base pairs) and longer ones. Even though ctDNA is usually less than 1% of the total cfDNA in the blood, it carries important changes in genes (like EGFR or TP53), which help doctors find cancer through blood tests.
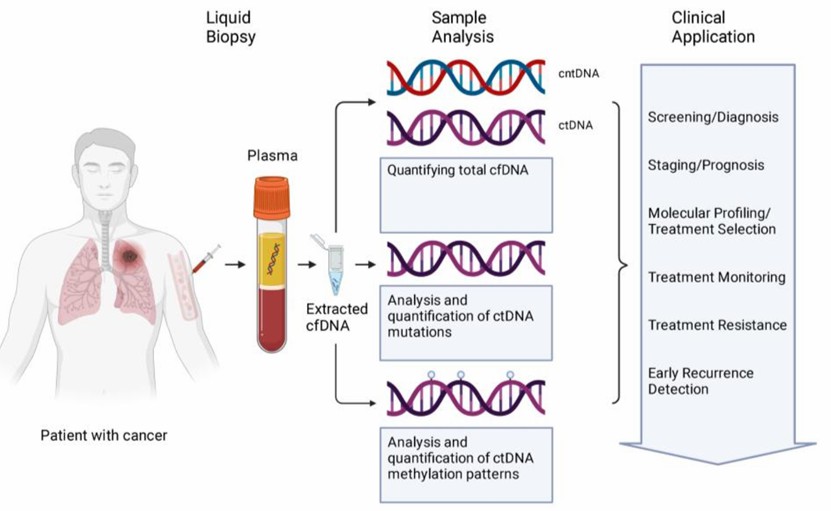
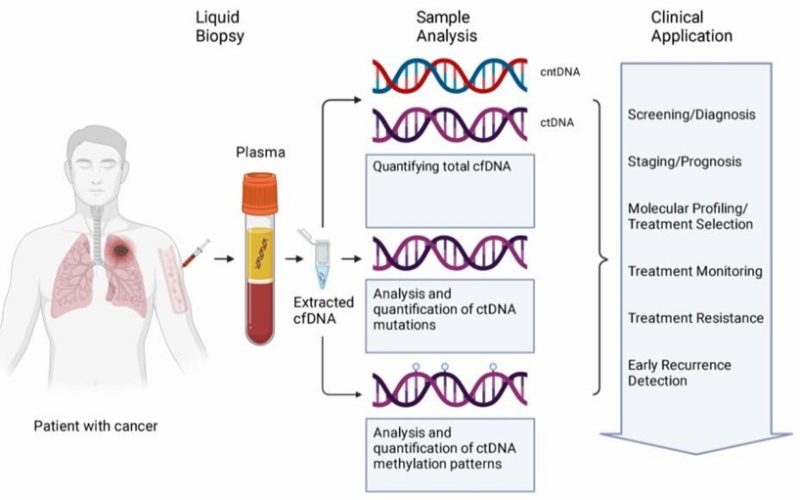
Figure 1. cfDNA from liquid biopsy samples are analyzed in several ways.( Jonathan Dao, et. al,2023)
| Feature | cfDNA | ctDNA |
|---|---|---|
| Source | Apoptotic/necrotic normal cells | Tumor cells (via necrosis, apoptosis, or secretion) |
| Fragment Size | Predominantly 166 bp (nucleosomal) | Shorter (<150 bp) + longer fragments |
| Concentration | 1-100 ng/mL plasma (healthy) | Often <1% of total cfDNA |
| Mutations | Wild-type | Tumor-specific (e.g., EGFR, TP53) |
- cfDNA reflects general cellular turnover.
- ctDNA carries cancer-specific alterations, enabling liquid biopsies.
2. Extraction Methods: Tailoring to Your Needs
The choice of extraction method for cfDNA and ctDNA depends on sample type, downstream applications, and target fragment size. Pre-analytical handling is critical to ensure high-quality yields, particularly for rare ctDNA variants.
2.1. Pre-Analytical Considerations
Blood Collection:
- cfDNA: EDTA tubes (process within 2-4 hours).
- ctDNA: Streck tubes (stable for 7 days at room temperature).
Plasma Prep: Double-centrifuge (1,600 × g → 16,000 × g) to remove cells.
2.2. Comparison of Extraction Techniques
| Method | Best for cfDNA? | Best for ctDNA? | Why? |
|---|---|---|---|
| Silica Columns | (QIAamp CNA Kit) | (loses short fragments) | High purity but misses small ctDNA |
| Magnetic Beads | (Good recovery) | (MagMAX, Dynabeads) | Better recovery of <150 bp fragments |
| Phenol-Chloroform | (low yield) | (high background) | Outdated for low-abundance targets |
Pro Tip: For ctDNA, add carrier RNA to improve recovery of rare fragments.
3. Detection & Analysis: Sensitivity Matters
The choice of detection methods for cfDNA and ctDNA must account for their distinct biological and clinical requirements. CfDNA analysis typically employs qPCR (ALU repeats) for quantification due to its cost-effectiveness and reliability in measuring total circulating DNA. In contrast, ctDNA demands ultra-sensitive techniques like ddPCR to detect rare tumor-derived mutations at frequencies as low as 0.01%.
For comprehensive profiling, whole-genome/exome sequencing (WGS/WES) provides broad coverage of cfDNA, making it suitable for non-cancer applications (e.g., NIPT, transplant monitoring). However, targeted NGS panels (e.g., Guardant360) are preferred for ctDNA, as they enhance sensitivity for cancer-specific variants while reducing costs.
Size analysis further differentiates the two:
- cfDNA: Standard Bioanalyzer assessment confirms the expected ~166 bp nucleosomal pattern.
- ctDNA: Fragmentomics (DELFI score) leverages aberrant fragmentation profiles to improve tumor detection, particularly in early-stage disease.
| Application | cfDNA Tools | ctDNA Tools |
|---|---|---|
| Quantification | qPCR (ALU repeats) | ddPCR (for rare mutations) |
| Profiling | WGS/WES | Targeted NGS panels (e.g., Guardant360) |
| Size Analysis | Bioanalyzer | Fragmentomics (DELFI score) |
3.1 Case Study: Non-Invasive Early-Stage Lung Cancer Diagnosis via ctDNA Methylation Profiling (Liang et al., 2019)
Testing ctDNA (cancer DNA in the blood) for changes in DNA marks can help doctors find early lung cancer. In a study with 230 lung nodules (small growths in the lung), this test was very accurate, with 92.7% sensitivity and 92.8% specificity when tested on tissue.
When they tried the same test on blood plasma, it still worked well. The 9-marker panel could find 79.5% of cancers and had a 85.2% accuracy in telling apart cancer from non-cancer growths. For early-stage lung cancer (stage IB), it was even better, with 85.7% accuracy. This test also did a great job in showing 93.2% accuracy in healthy people, helping avoid false alarms.
The test even found 75% of the cancers in their very early stages (stage IA), which is a big deal because it’s hard to catch cancer so early. It could also help doctors avoid 85% of unnecessary biopsies (needle tests) in cases that aren’t cancer, making it way better than traditional scans at reducing false positives.
Why it matters: This is the first time ctDNA testing has shown it can find lung cancer early with just a blood test. This research is a big step toward developing better liquid biopsy tools for doctors.
4. Clinical Applications: Where Each Shines
The distinct biological properties of cfDNA and circulating tumor ctDNA determine their unique clinical utilities. While cfDNA is derived from normal cell turnover and can originate from various tissues, ctDNA is specifically shed by tumor cells, carrying cancer-associated mutations and epigenetic alterations. This fundamental difference dictates their applications in non-cancer diagnostics versus oncology.
4.1 Non-Cancer Uses of cfDNA
cfDNA has emerged as a powerful tool in non-oncological diagnostics due to its minimally invasive nature and broad tissue representation. Key applications include:
Invasive Prenatal Testing (NIPT)
- Fetal Chromosome Abnormalities: cfDNA from the placenta (called cell-free fetal DNA, or cffDNA) is used to check for problems with the baby’s chromosomes, like Down Syndrome (Trisomy 21), Edwards Syndrome (Trisomy 18), and Patau Syndrome (Trisomy 13).
- How Well It Works: For Down Syndrome (Trisomy 21), NIPT is very accurate, with over 99% sensitivity and specificity. This means doctors can find out if there’s an issue without needing risky tests like amniocentesis (a needle test).
- More Uses: New technology also lets doctors find even smaller genetic changes, determine the baby’s sex, and check for single-gene disorders.
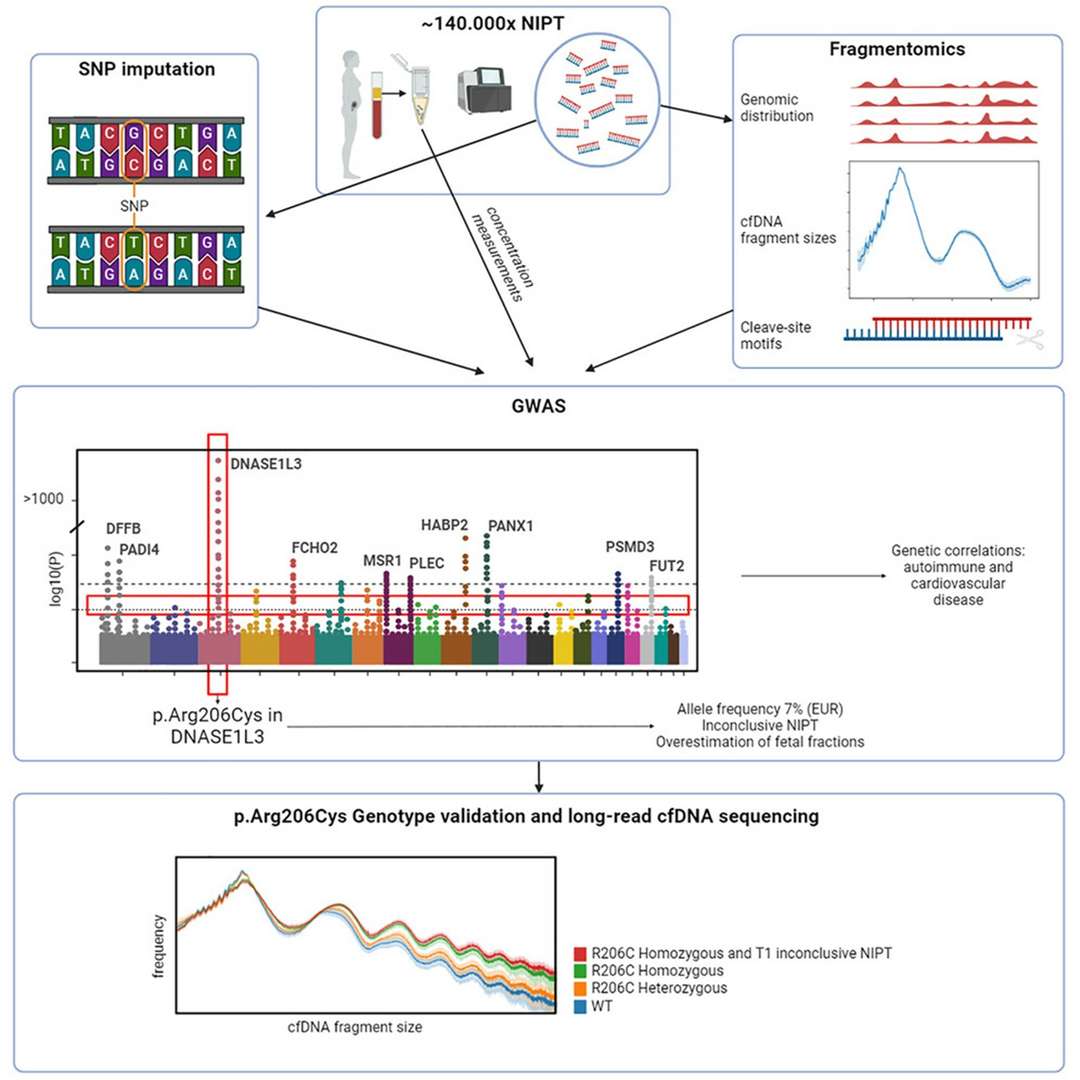
Figure 2.Common genetic variants and cfDNA characteristics.( Jasper Linthorst, et. al,2024)
Transplant Rejection Monitoring
Donor-Derived cfDNA (dd-cfDNA): After organ transplantation, elevated levels of donor-derived cfDNA in the recipient’s blood correlate with allograft injury or rejection.
Diagnostic Accuracy: Studies report an AUC (Area Under the Curve) of 0.91, making it a reliable biomarker for early rejection detection, particularly in heart and kidney transplants.
Advantages Over Biopsy: Unlike invasive tissue biopsies, dd-cfDNA offers a real-time, dynamic assessment of graft health, enabling timely intervention.
Other Emerging Applications
Autoimmune Diseases: cfDNA methylation patterns may help monitor disease activity in lupus (SLE) and rheumatoid arthritis.
Sepsis & Trauma: Elevated cfDNA levels correlate with tissue damage severity, aiding in prognosis and therapeutic response assessment.
4.2. Oncology Applications of ctDNA
ctDNA, carrying tumor-specific mutations, has revolutionized liquid biopsy in cancer management. Its applications span:
Early Detection & Screening
Multi-Cancer Early Detection (MCED): Panels targeting methylation patterns and somatic mutations can identify multiple cancer types (e.g., lung, breast, colorectal) at early stages.
High-Risk Populations: Useful for Lynch syndrome, BRCA carriers, and heavy smokers to detect malignancies before clinical symptoms arise.
Treatment Selection & Personalized Therapy
Identifying Targetable Mutations: ctDNA analysis detects EGFR, KRAS, BRAF, and PIK3CA mutations, guiding tyrosine kinase inhibitor (TKI) or immunotherapy choices.
Resistance Monitoring: Emergence of EGFR T790M or ALK resistance mutations in NSCLC can prompt therapy adjustments.
Minimal Residual Disease (MRD) & Recurrence Monitoring
Post-Surgical Surveillance: ctDNA positivity after tumor resection predicts relapse months before imaging (e.g., in colorectal and breast cancers).
Dynamic Risk Stratification: Serial ctDNA tracking helps distinguish patients needing adjuvant therapy from those with low recurrence risk.
Therapeutic Response Assessment
Real-Time Efficacy Monitoring: Declining ctDNA levels post-treatment correlate with tumor regression, while rising levels indicate progression or resistance.
Clinical Trials: ctDNA is increasingly used as a surrogate endpoint in trials to accelerate drug development.
| Use Case | Example |
|---|---|
| Early Detection | GRAIL’s Galleri test (50+ cancers) |
| Treatment Monitoring | EGFR T790M in NSCLC (Guardant360 CDx) |
| MRD Detection | Signatera (tumor-informed assays) |
Real-World Impact:
The BESPOKE-CRC trial demonstrated ctDNA’s clinical utility, where ctDNA-guided therapy reduced unnecessary chemotherapy by 48% in colorectal cancer patients, avoiding overtreatment while maintaining efficacy.
5. Troubleshooting Guide
Effective cfDNA/ctDNA testing faces three big challenges: getting enough DNA, contamination from other DNA, and problems with the testing process.
- Low DNA Amounts: To get more cfDNA, use at least 4 mL of plasma. Bead-based extraction helps capture rare ctDNA fragments.
- Contamination by gDNA: To remove extra gDNA, use DNase treatment for cfDNA. For ctDNA, using two rounds of spinning (1,600g → 16,000g) and sorting the DNA by size works best.
- Testing Problems (PCR Inhibition): For cfDNA, simply diluting the sample (1:5) often works. But ctDNA needs special steps to remove any inhibitors because it’s present in very low amounts.
One important thing to remember is hemolysis, which is when blood breaks down and makes the plasma look pink. This makes it harder to see ctDNA signals.
To avoid this, use blunt needles when collecting blood, check the quality of the plasma with special tests (like A414/A375 ratios), and use control samples to make sure the extraction is working properly.
By using these methods, scientists can find cfDNA and ctDNA more easily. This helps with important tests like cancer monitoring and non-invasive prenatal testing.
Common Challenges
| Issue | cfDNA Solution | ctDNA Solution |
|---|---|---|
| Low Yield | Increase plasma volume (≥4 mL) | Use bead-based extraction |
| gDNA Contamination | DNase treatment post-extraction | Double-centrifuge + size selection |
| PCR Inhibition | Dilute sample 1:5 | Add inhibitor removal step |
Critical Note: Hemolysis increases wild-type DNA, masking ctDNA signals-always check plasma for pink discoloration!
Conclusion
The comprehensive comparative analysis of cfDNA and ctDNA elucidates their distinct yet complementary roles in advancing modern diagnostic paradigms and therapeutic monitoring strategies. These circulating nucleic acids offer unique windows into human health and disease states as two critical components of liquid biopsy technologies. cfDNA, with its broad representation of cellular turnover dynamics, serves as a powerful systemic biomarker, providing indispensable clinical utility in three key areas: (1) NIPT, where it has revolutionized fetal genetic screening with >99% detection rates for common trisomies; (2) solid organ transplant surveillance, enabling sensitive detection of graft rejection through donor-derived DNA quantification; and (3) inflammatory disease monitoring, offering real-time assessment of disease activity in conditions like lupus and rheumatoid arthritis.
Conversely, ctDNA has emerged as a transformative tool in oncology, with three principal clinical applications demonstrating its unique value: (1) early cancer detection, particularly for hard-to-diagnose malignancies like pancreatic and ovarian cancers, where recent studies (e.g., DETECT-A trial) have shown detection rates exceeding 70% for stage I/II disease; (2) therapy selection, where ctDNA profiling identifies actionable mutations with 90-95% concordance with tissue biopsies (as demonstrated in the TARGET trial); and (3) minimal residual disease monitoring, with tumor-informed ctDNA assays predicting recurrence 6-9 months before radiographic evidence (per TRACERx data).
The integration of these analytes in clinical practice requires careful consideration of their distinct biological characteristics. While cfDNA analysis benefits from standardized isolation protocols and relatively abundant yields (typically 5-100 ng/mL plasma), ctDNA detection demands ultra-sensitive approaches due to its low fractional abundance (<0.1% in early-stage cancers). Emerging solutions to these challenges include: (1) novel preservation tubes (e.g., Streck cfDNA BCT) that maintain sample integrity, (2) optimized extraction methods favoring magnetic beads over columns for short-fragment recovery, and (3) advanced detection platforms combining ultra-deep sequencing (100,000× coverage) with error-correction algorithms.
Future directions in the field point toward three transformative developments: (1) multi-analyte liquid biopsy panels integrating cfDNA, ctDNA, methylation, and fragmentomics signatures; (2) point-of-care microfluidic platforms enabling rapid turnaround; and (3) artificial intelligence-driven interpretation algorithms. As these technologies mature, they promise to establish liquid biopsy as a cornerstone of precision medicine, with projected market growth to $28 billion by 2028 according to recent industry analyses.
For optimal clinical implementation, we recommend: (1) adopting consensus pre-analytical standards (e.g., CLSI guidelines) to minimize variability, (2) validating assay performance in intended-use populations, and (3) developing interdisciplinary workflows combining laboratory medicine, bioinformatics, and clinical expertise. By strategically leveraging the complementary strengths of cfDNA and ctDNA, the medical community can realize the full potential of liquid biopsies to transform disease detection, monitoring, and management across diverse clinical contexts.
Read More:
Certified Hair Implant Clinics
References:
- Dao, J., Conway, et, al., (2023). Using cfDNA and ctDNA as Oncologic Markers: A Path to Clinical Validation. International journal of molecular sciences, 24(17), 13219. https://doi.org/10.3390/ijms241713219